ٱلْحَمْدُ لِلَّٰهِ I have had the privilege of doing research both in my undergraduate and postgraduate years. I thank my supervisors and collaborators for bringing out the best in me. I will try to explain what I did in layman terms. Abstract and paper are also included. I did my Bachelors from the Faculty of Science at the University of Colombo. I was in the Computational Physics Honors program. Currently I am at the end of my PhD at Indiana University-Purdue University Indianapolis (IUPUI). I’m at the Physics Department in the School of Science. I’m not sure what my next step will be. I will let you know إِنْ شَاءَ ٱللَّٰهُ
PhD (IUPUI, USA – present)
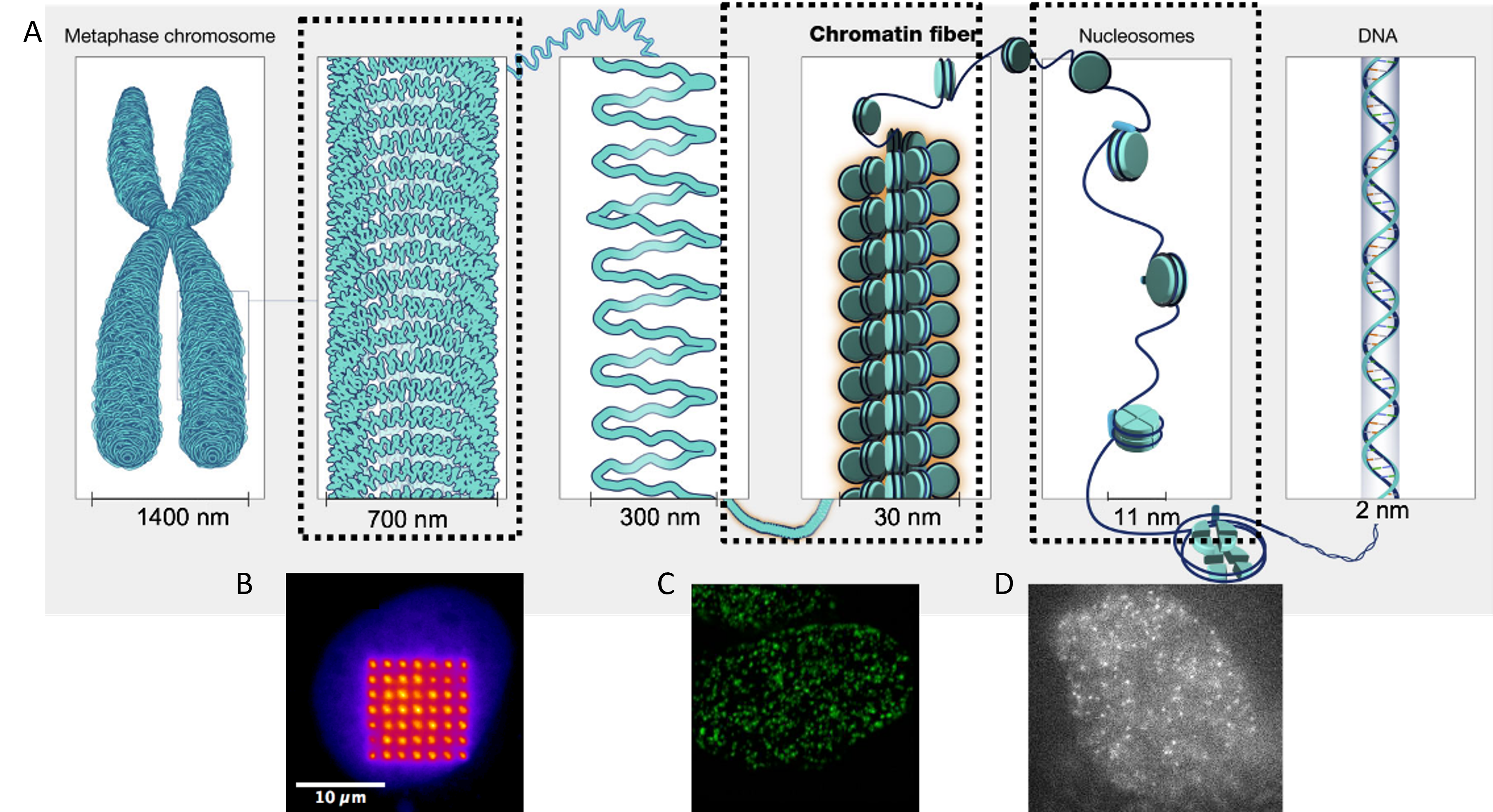
DNA contains the genetic code that makes us who we are and is packed in the nucleus of our cells as chromosomes. There’s an intermediate structure called chromatin where DNA is wrapped around balls of histones. Thanks to advancements in microscopy, we now know that they move rapidly. For Biophysicists it has been an interesting problem to understand how chromatin moves. I know, we like to come up with models that predict the behavior of nature. How chromatin responds to drugs, or DNA damage, and developing tools to sensitively detect the behavior of chromatin is my objective.
Masters (IUPUI, USA – 2021)
I capture movies of fast-moving molecules of DNA in human cells under a microscope. One of the biggest bottlenecks was when you increase the frame rate of the camera, the signal of the image reduces. Artificial Intelligence (AI) had it’s newest family member, known as Deep Learning (DL). We have seen DL restoring low signal images and we thought, can we use that on our data?
My role was to:
- Learn DL from scratch and look at existing DL networks
- Train those networks with our data: Synthetic and real microscopic cell images
- Restore low signal movies using those networks
- Is the restoration reliable? Validate those outputs

It turns out, DL does show some promise. However, it can lie confidently too. This was my first paper from my PhD and I am so thankful to the amazing Chromatin group that I was a part of. My supervisor was Dr. Jing Liu.
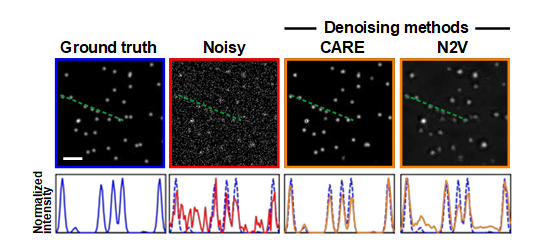
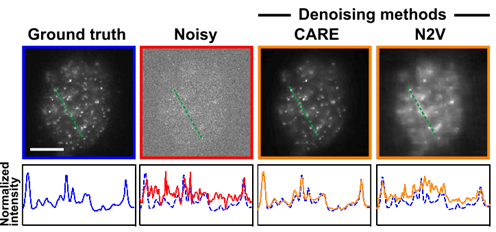
Here’s our paper:
Kefer, Paul, et al. “Performance of deep learning restoration methods for the extraction of particle dynamics in noisy microscopy image sequences.” Molecular Biology of the Cell 32.9 (2021): 903-914.
Bachelors (University of Colombo, Sri Lanka – 2016)
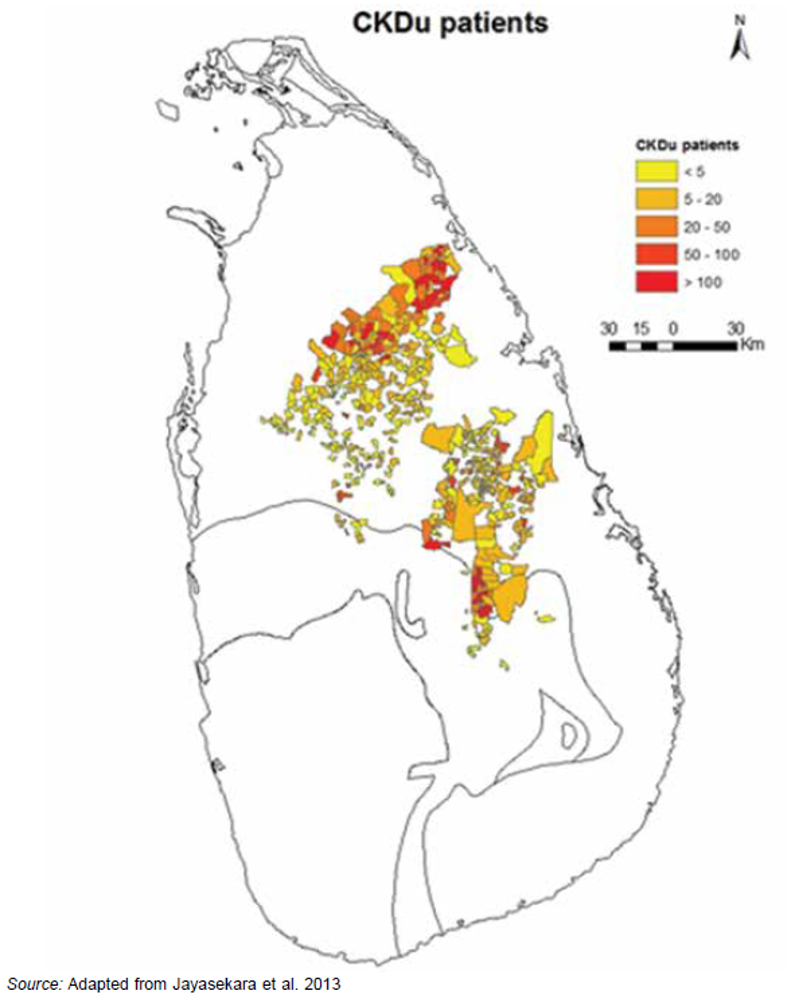
Chronic kidney diseases of unknown etiology (CKDu) is prevalent in the agricultural provinces in Sri Lanka. The root cause is not confirmed. Under the supervision of Dr. Janaka Wansapura, our lab wanted to explore the possibility of creating an early detection mechanism to reduce the risk of kidney loss among patients. To get ultrasound kidney images and understand them I had the help of Dr. Aruna Pallewatte from the National Hospital of Sri Lanka (Department of Nephrology)
My role was to:
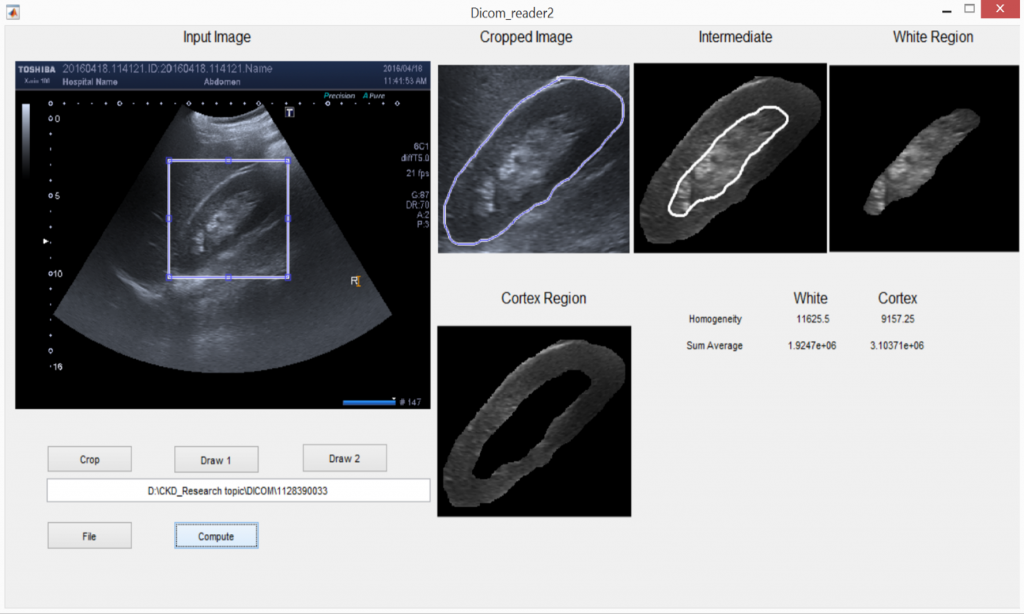
- Build a computational pipeline on MATLAB which can read ultrasound images of kidneys
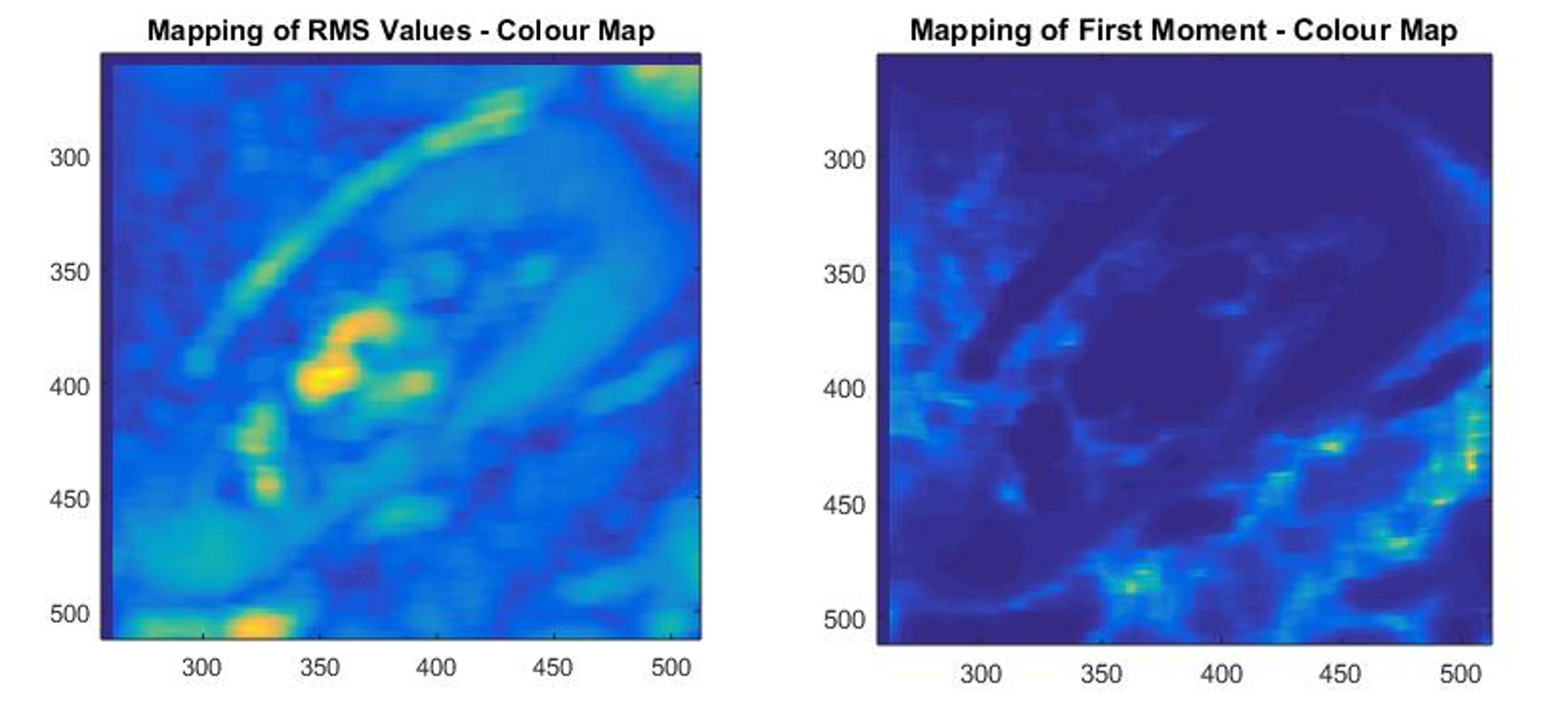
- Extract different statistical parameters from the pixel values on different regions in the kidney
- Can these parameters distinguish between sick and healthy kidneys?
We did come across interesting parameters and you can learn more from my paper. Many thanks to my supervisor and Doctor Pallewatte for all the help!
Iqbal, Fadil, Aruna S. Pallewatte, and Janaka P. Wansapura. “Texture analysis of ultrasound images of chronic kidney disease.” 2017 Seventeenth International Conference on Advances in ICT for Emerging Regions (ICTer). IEEE, 2017.